
OpenProt is the first database to hold a polycistronic annotation model for eukaryotic transcriptomes. Here, we highlight the use of OpenProt databases for MS-based proteomics. Although most of these discoveries have been serendipitous, they demonstrate the caveats of current genome annotations and the polycistronic nature of eukaryotic genes 8. These novel proteins are found encoded in allegedly non-coding RNAs, in the 5' or 3' untranslated regions (UTR) of mRNAs, or overlapping the canonical coding sequence (cCDS) in an alternative frame. An increasing number of studies challenge the current annotation model and report discoveries of unannotated functional ORFs in eukaryotic genomes 8, 11, 12, 13, 14. However, genome annotations hold arbitrary criteria for ORF annotation, such as a minimum length of 100 codons and a single ORF per transcript 9, 10. This method relies on current genome annotations to generate a reference protein sequence database that outlines the scope of possibilities 6, 7, 8. Over the past decades, mass spectrometry (MS-)based proteomics has become the golden technique to decipher proteomes of eukaryotic cells 1, 2, 3, 4, 5. Overall, OpenProt is a freely available tool that will foster proteomic discoveries. However, with appropriate false discovery rate (FDR) settings or the use of a restricted OpenProt database, users will gain a more realistic view of the proteomic landscape. The size of OpenProt database (all predicted proteins) is substantial and need be taken in account for the analysis. Using OpenProt database for proteomic experiments enables novel proteins discovery and highlights the polycistronic nature of eukaryotic genes. OpenProt is freely accessible and offers custom downloads of protein sequences across 10 species. OpenProt is the first database that enforces a polycistronic model for eukaryotic genomes, allowing annotation of multiple ORFs per transcript.
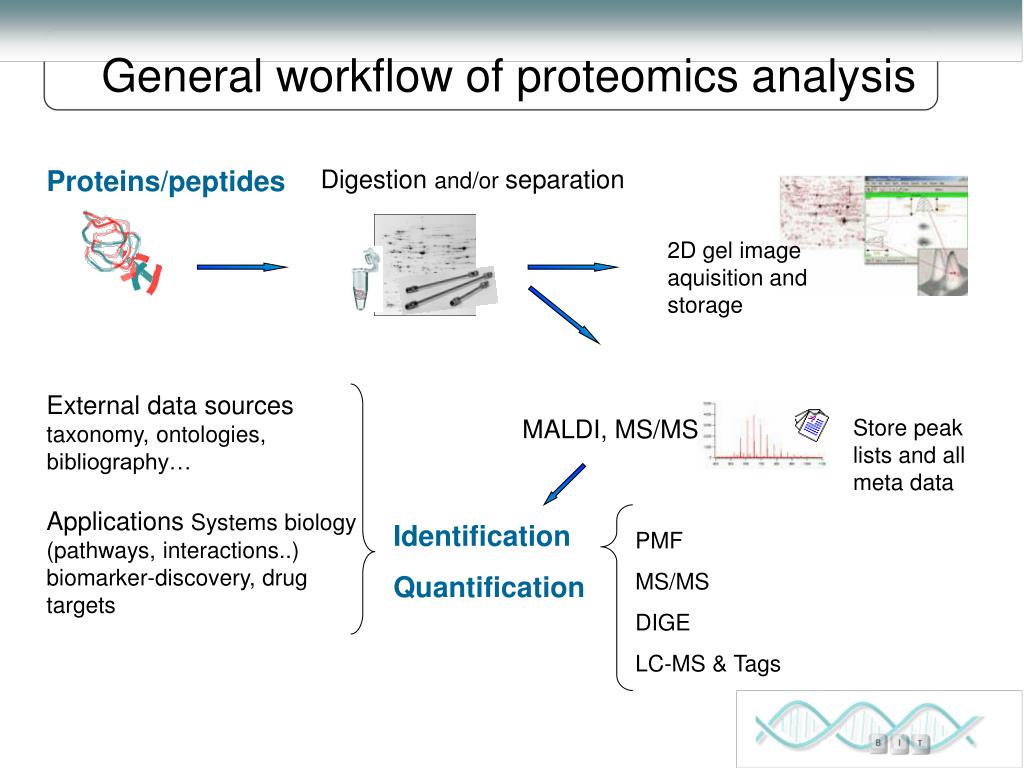
These novel proteins were found encoded either within non-coding RNAs, 5' or 3' untranslated regions (UTRs) of mRNAs, or overlapping a known coding sequence (CDS) in an alternative ORF. However, a growing number of studies report expression of proteins from allegedly non-coding regions, challenging the accuracy of current genome annotations. Traditional models of open reading frame (ORF) annotation impose two arbitrary criteria: a minimum length of 100 codons and a single ORF per transcript. Genome annotation is central to today's proteomic research as it draws the outlines of the proteomic landscape.


 0 kommentar(er)
0 kommentar(er)
